Go to the directory where your files are stored (VEC, VC7, MAT files or others), and type
v = loadvec('B00001.VC7');v = loadvec('*.VC7');To display the fields, typev = loadvec(1:10);
There are several shortcuts: For instance, this will display a movie of all the files in the current directory:showf(v);
showf('*.VC7');To get information about the function loadvec, type
(open the help browser with the help for this function), ordoc loadvec
(display the text of the help in the command window). If you don't know what you look for, have a look to the Function by category section from the PIVMat main page:help loadvec
docpivmat
You should be familiar with the basic Matlab syntax, especially the use of colons (:) to index arrays, and the use of structures and structure arrays.
Use filterf or bwfilterf, eg:
showf(filterf(v,2));
First load your vector field with
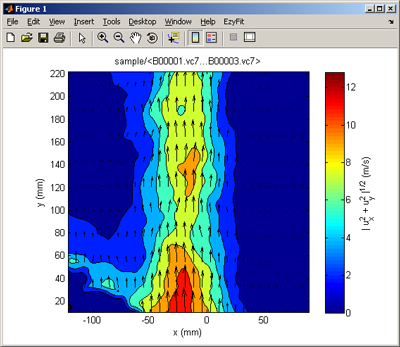
v = loadvec('B00001.VC7');Another way to do this is to create a scalar field curl,showf(v,'rot');
and then to display it:curl = vec2scal(v,'rot');
Before computing the vorticity field, you will probably want to filter your vector field:showf(curl);
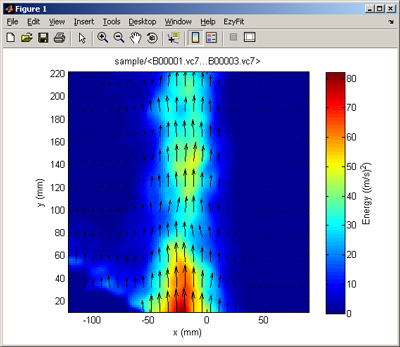
Many other vector-to-scalar conversions (divergence, kinetic energy, velocity derivatives...) are available from vec2scal.showf(filterf(v,1),'rot');
You may load all the vector fields and display only the ones you want:
v = loadvec('*.VC7');If you want to refer to a range of fields, n may be an array, like [1 3 8], or 1:10 (type doc colon to learn more about Matlab's colon (:)). So, to display only the first 10 fields, type
If you want to skip one field every 2, typeshowf(v(1:10));
You may also only load the fields you want, by using wildcards (*) and/or brackets []. In order to load only the files B00001.VC7 to B00010.VC7, typeshowf(v(1:2:10));
v = loadvec('B[1:10].VC7');(in this case, the 10 first files are loaded in the alphabetically-ordered set of files of the current directory).v = loadvec(1:10);
See the page Importing data into PIVMat to learn more.
Yes, loadvec accepts IMX/IM7 files; they will be considered as normal scalar fields, ie: use showf to display them. You may use colormap gray to use a black-and-white color map, which is usually more suitable for images.
Yes. Simply use loadvec (see also loadpivtxt). However, if you work with DaVis, it is better to import directly the VC7 files: it is faster, and these files contain many additional informations.
A vector field is a bit confusing when too many vectors are present in a field. By default, showf displays at most 32 vectors in each direction, whatever the vector field resolution. This can be changed using the 'Spacing' option of showf, eg:
You may also display specify different spacing in the x- and y-direction:showf(v,'norm','spacing',2);
showf(filterf(v,1.5),'ken','spacing',[4 12]);
The ReadIMX package provided by LaVision allows to import a (single) VC7/IMX/IM7 file from Davis into Matlab, and to display it. That's all.
The PIVMat toolbox is based on the function readimx from the ReadIMX package, but offers much more. Note that the ReadIMX package has to be installed separately for the PIVMat toolbox to work correctly (see the Installation page).
PIVMat is compatible with stereo (3-components) velocity fields only for DaVis files.